Successfully Added to Cart
Customers also bought...
-
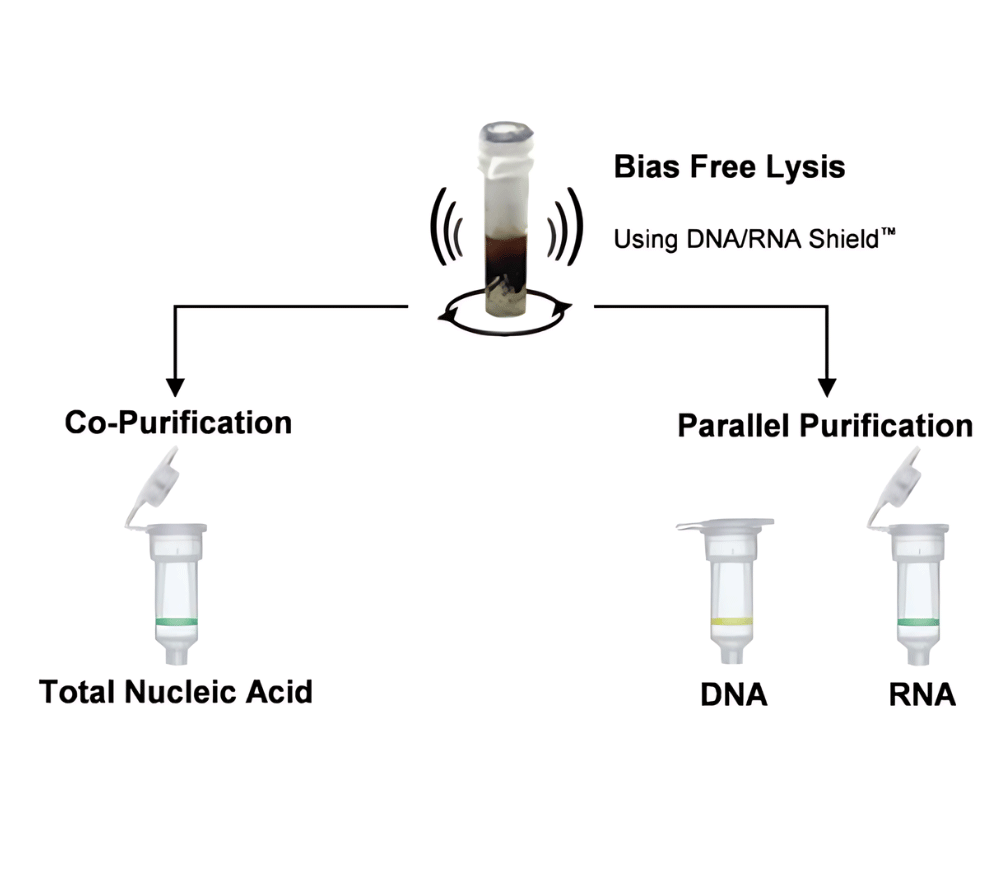
 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
ZymoBIOMICS Microbial Community Standard II (Log Distribution)
| Cat # | Name | Size | Price | Quantity |
|---|
Highlights
- Log abundance distribution: assess detection limit over a broad range (102 to 108 cells)
- Accurate composition: assess the accuracy of microbiome measurements
- Microbiomics QC: ideal for quality control of microbiome measurements
Documents
Product Description
Technical Specifications
| Applicable For | NGS, microbiomics, metagenomics. |
|---|---|
| Processing Volume | 75 µl per preparation. |
| Purity | Contains < 0.01% foreign microbial DNA. |
| Sample Source | Eight bacteria (3 Gram-negative and 5 Gram-positive) and 2 yeasts. |
| Yield | Approximately 220 ng DNA per preparation. |
Resources
Q1: Which standard should I use and how do I use it?
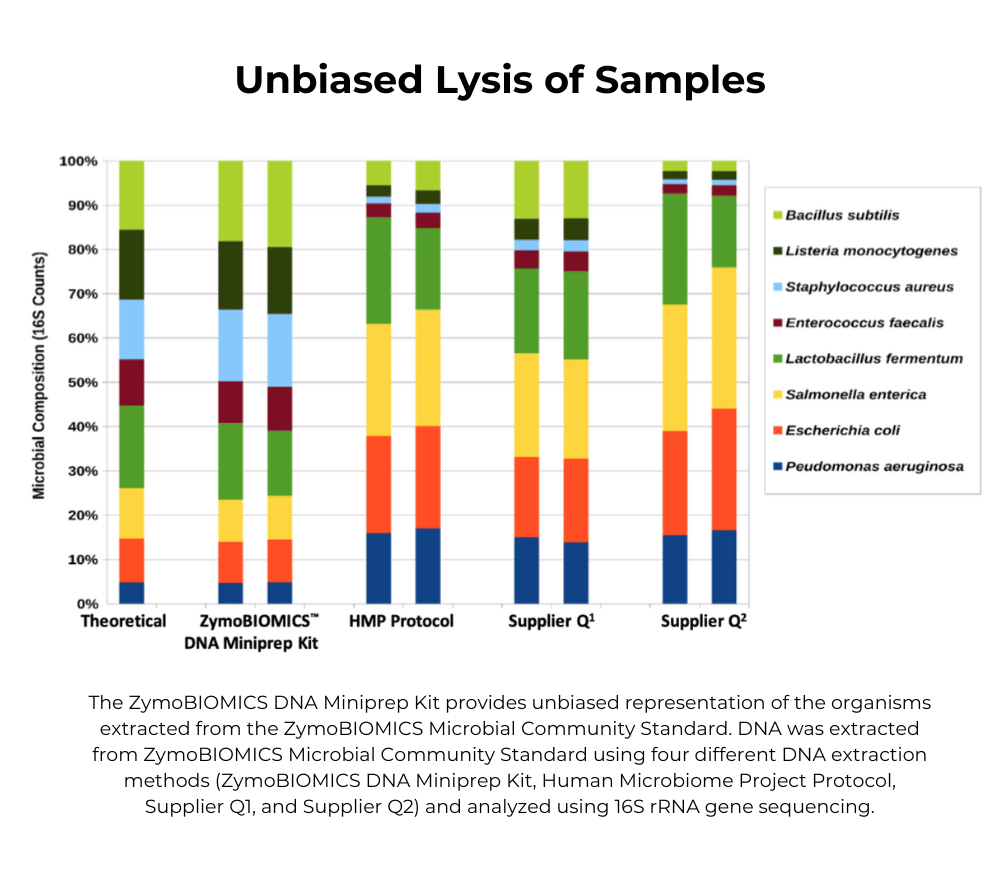
We recommend working backwards from the analysis. First optimize your library prep with the ZymoBIOMICS Microbial Community DNA Standard (D6305, D6311) to assess bias in PCR, sequencing, bioinformatics, etc. Once your results have low/no bias when compared to the theoretical composition, then use the whole cell ZymoBIOMICS Microbial Community Standard (D6300, D6310) to assess bias in lysis efficiency.
Q2: Is the standard supposed to be spiked into a sample?
No, the Microbial Community Standard is designed to assess the efficiency of the lysis method and is intended to be run in parallel with your samples. If all the organisms can be detected at/near the theoretical abundance, you can be confident your extraction method is unbiased.
Q3: Is the standard compatible with Qiagen kits?
The chemistry of the Qiagen Kits (QIAamp Powerfecal, DNeasy Powersoil) and the storage solution the Microbial Community Standard is stored in (DNA/RNA Shield) are not completely compatible. Instead, less input volume should be used (25 ul).
Q4: Where can I find your reference files for the genomes of your Standard strains (D6300, D6305, and D6306)?
You can find the reference genome and 16S/18S sequences here: ZymoBIOMICS.STD.refseq.v2.zip.
Q5: Why do my abundance results not match the theoretical composition?
This may indicate an issue with the lysis method, which can be biased toward gram negative bacteria. See the Optimized Lysis Protocols under Documents for a table of bead beating devices and protocols validated by Zymo Research.
Devices found to underrepresent tough-to-lyse organisms under all tested conditions:
- Retsch Mixer Mills
- Tissues Lyzers
- MP Fast Prep 96
Q6: What database was used to generate the data for the standards on the data sheet?
We use an in-house curated database to generate the data for the standards.
Q7: Is the raw sequencing data for the standards available?
Please contact Technical Support at tech@zymoresearch.com for raw sequencing data.
Q8: What is the expected DNA fragment length in the Microbial Community DNA Standard? (D6305/D6306)
The expected DNA fragment size is <15kb.
Q9: How come I am unable to detect certain organisms in the standard?
This could indicate bias in the workflow, such as inefficient lysis, library prep or bioinformatics analysis.
Related Products
D4300, D4300T, D4304
ZymoBIOMICS DNA Miniprep Kit
Efficient microbial DNA purification from low sample inputs for highly accurate microbiome and metagenome analyses.
R2002
ZymoBIOMICS DNA/RNA Miniprep Kit
The ZymoBIOMICS DNA/RNA Miniprep Kit is designed for purifying DNA and RNA from a wide array of sample inputs (e.g. feces, soil, plant, water, and biofilms) ...
D6421-PS1, D6421-PS2, D6421-PS3...
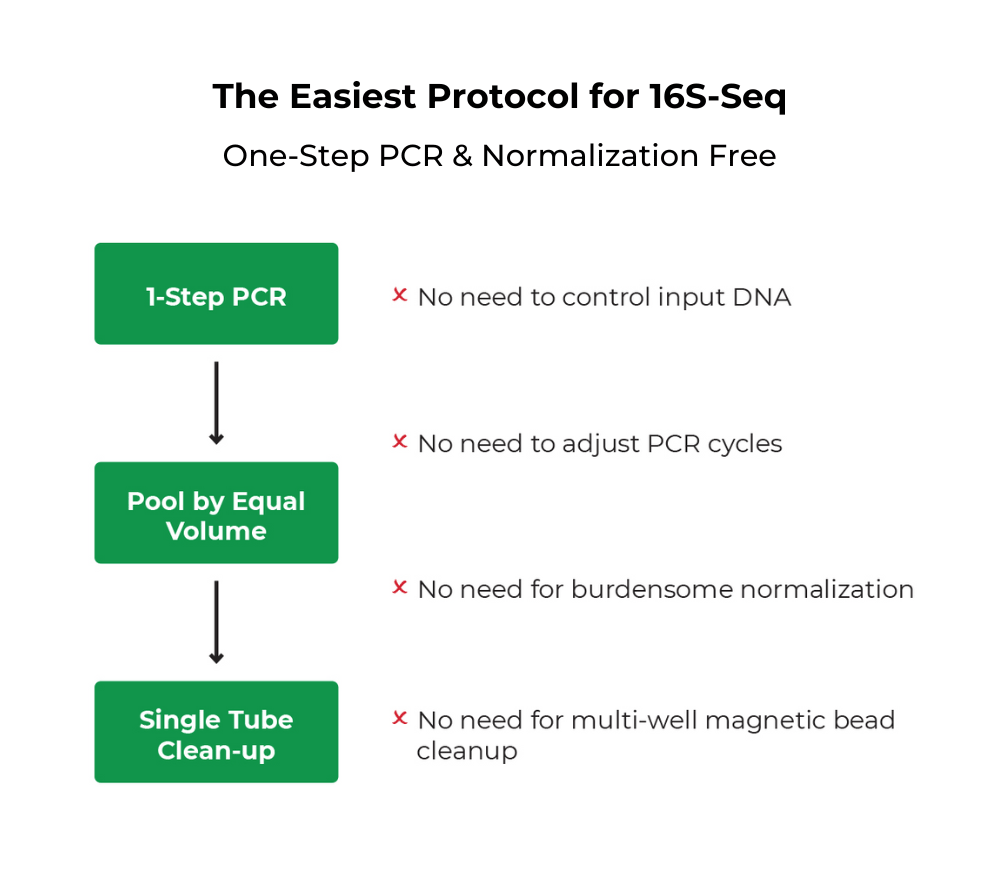
Quick-16S Plus NGS Library Prep Kit (V3-V4, UDI)
The Quick-16S Plus NGS Library Prep Kit (V3-V4) is the fastest and simplest NGS library prep kit targeting the 16S rRNA gene for microbiome profile analysis.
Need help? Contact Us