How to Set up a Sequencing Run on the NextSeq 2000 Sequencing System
For libraries prepared using:
Once the libraries are prepared, you can proceed with the actual sequencing step, and the Illumina NextSeq 1000/2000 sequencing systems have become increasingly utilized over recent years.
The NextSeq 1000/2000 sequencing systems utilize patterned flow cells and provide output scalability and sequencing flexibility. They also come with an onboard implementation of the DRAGEN (Dynamic Read Analysis for GENomics) Bio-iT Platform that further streamlines sequencing data demultiplexing, transfer, and analysis.
Below is a step-by-step guide to appropriately set up a sequencing run on a NextSeq 2000 sequencer using libraries prepared with the Zymo-Seq RiboFree Total RNA Library Kit as an example to help researchers make the most of the NGS technologies.
Prerequisites
- Ensure that each sample being pooled and sequenced on the same lane of a sequencing run has been prepared using unique library barcodes.
- Zymo-Seq UDI Primers are provided as part of the Zymo-Seq RiboFree Total RNA Library Kits as either 1.5-mL tubes (R3000, UDI 1-12) or a 96-well plate (R3003, UDI 1-96). UDIs 1-12 in the tube format and the plate format contain identical UDIs. Be sure to take note of the UDI number being used for each sample during library preparation.
- All Zymo-Seq UDI Primer barcode sequences can be found in the Documents section of the Zymo-Seq UDI Primer Sets page or the Zymo-Seq RiboFree Total RNA Library Kit page. The barcodes are 8 bp long.
- When libraries are prepared with other kits from Zymo Research, be sure to look for the barcode sequences on the product page or the instruction manual accordingly.
- In this example, the NextSeq 2000 is compatible with a v2 sample sheet from Illumina’s run setup software. Confirm the sample sheet version applicable to your sequencer to ensure the appropriate barcode sequences and sample sheets are supplied. Also see more on NextSeq 1000/2000 sample sheets in this blog from Illumina.
Detailed Steps for Run Setup
- Log into the Illumina BaseSpace Sequence Hub with your user credentials.
-
On the home page, select the “Runs” Tab:

-
On the Runs page, click “New Run” dropdown menu on the right side of the page, and select “Run Planning”:
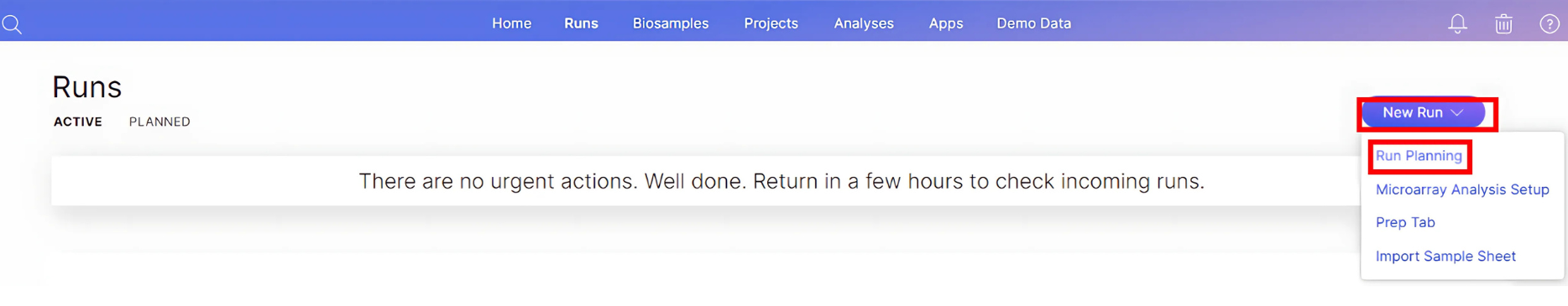
-
Fill out the “Create a Run” page with your run info:
- Run Name: The Run Name can contain 255 alphanumeric characters, dashes, underscores, periods, and spaces; and must start with an alphanumeric, a dash or an underscore.
- Run Description: This is optional.
- Instrument Platform: Select NextSeq 1000/2000.
- Secondary Analysis: Select Local if you will be using on-instrument DRAGEN pipelines or your own data analysis pipelines for secondary analysis of the sequencing data; select BaseSpace if you would like the data to be uploaded to BaseSpace and analyzed with DRAGEN pipelines in the BaseSpace Sequence Hub.
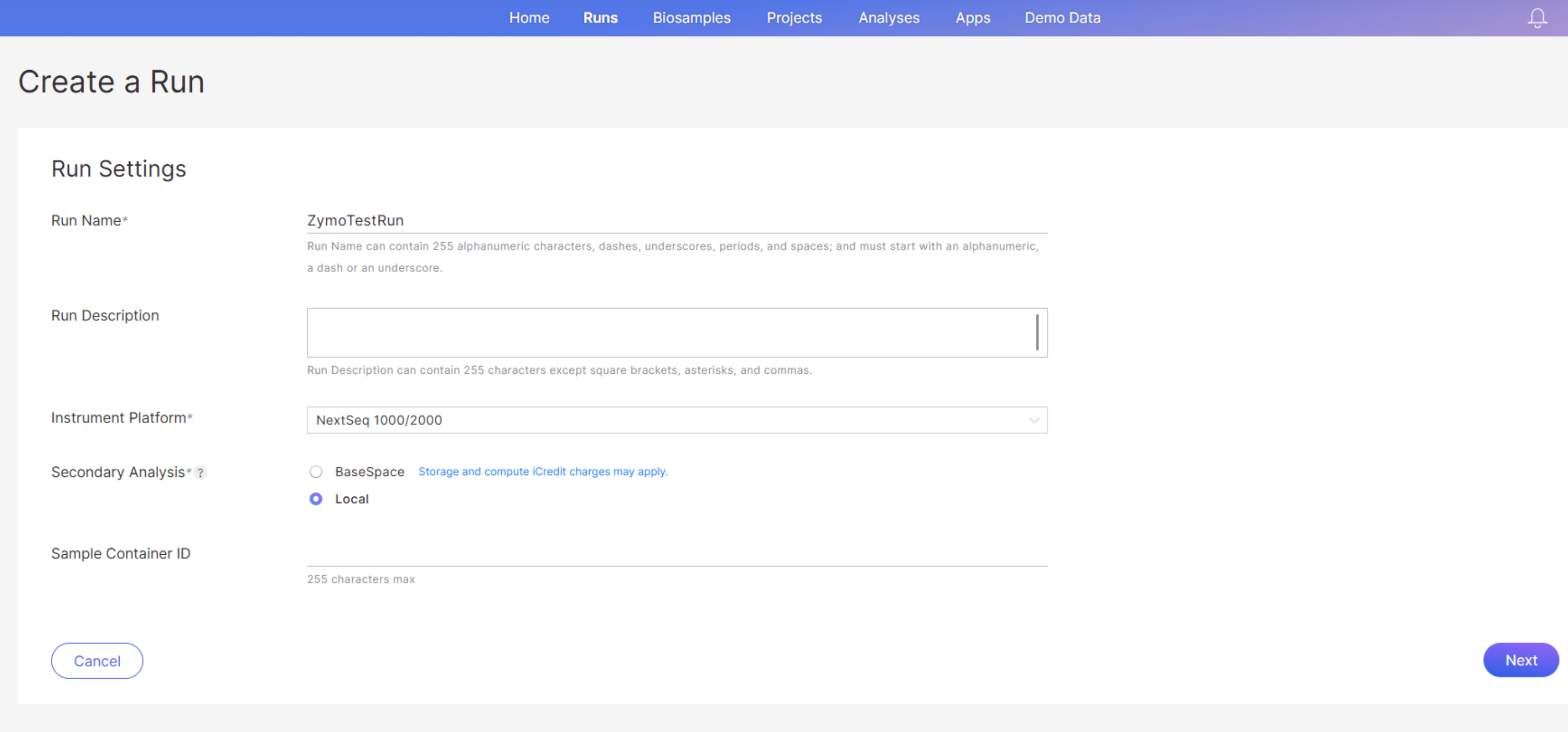
-
Click “Next”. On the “Configuration” page, select your analysis and library prep:
- Application: Select the secondary analysis you would like to be performed. Here “Illumina DRAGEN BCL Convert” is chosen to ensure the binary base call files are converted to FASTQ files that are requested for downstream bioinformatic analyses.
- Library Prep Kit: Select “Not Specified”
- Index Adapter Kit: Select “Not Specified”
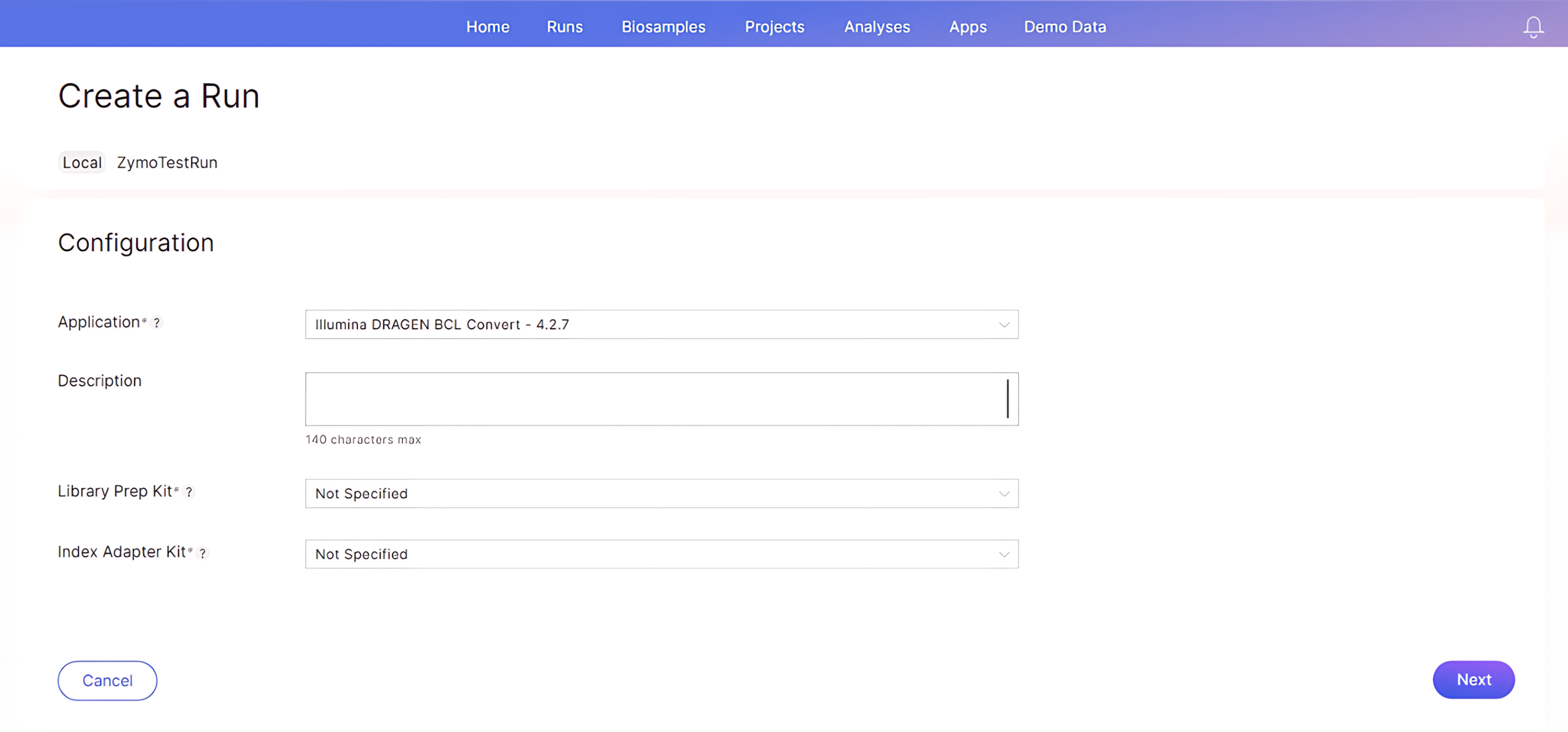
-
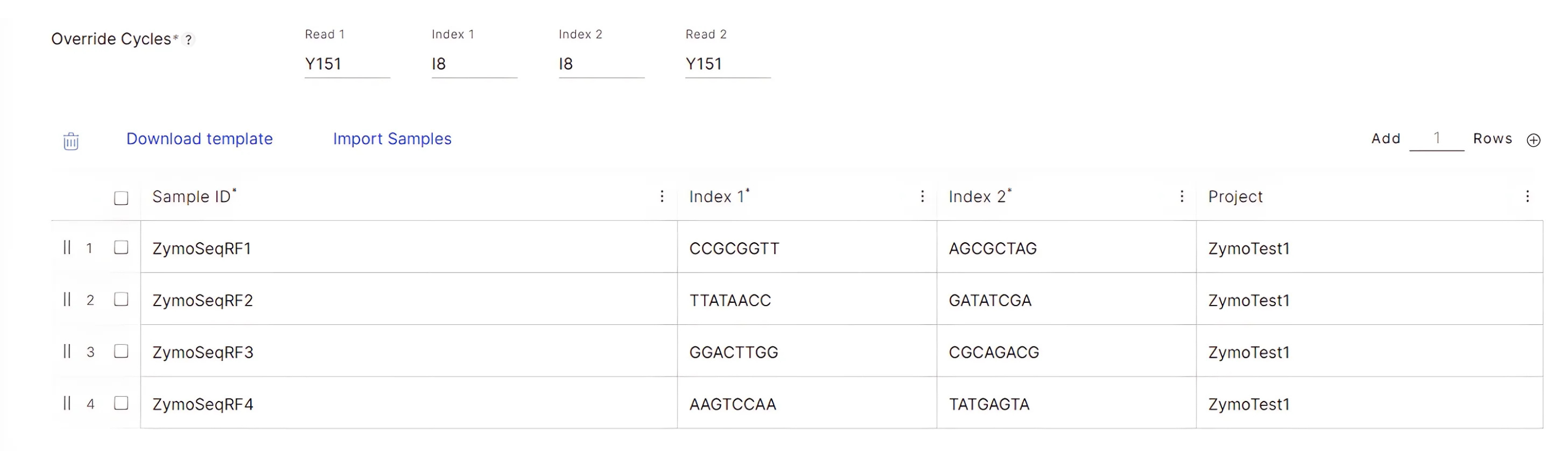
Click “Next”. On the following “Configuration…” page, input the run cycle parameters according to your libraries and the sequencing reagent kit. In this example we assume the use of a 300-cycle reagent kit for a 150 bp paired-end run to sequence the Zymo-Seq RiboFree libraries that have 8-bp Unique Dual Indexes:
- Index Reads: Select “2 Indexes”.
- Read Type: Select “Paired End”.
- Read Lengths: Input 151 for Read 1 and Read 2 read lengths. Input “8” for Index 1 and Index 2 read lengths.
- Override Cycles: This field will be automatically populated based on the parameters the user specified in the “Read Lengths” section. No modifications are required for this section.
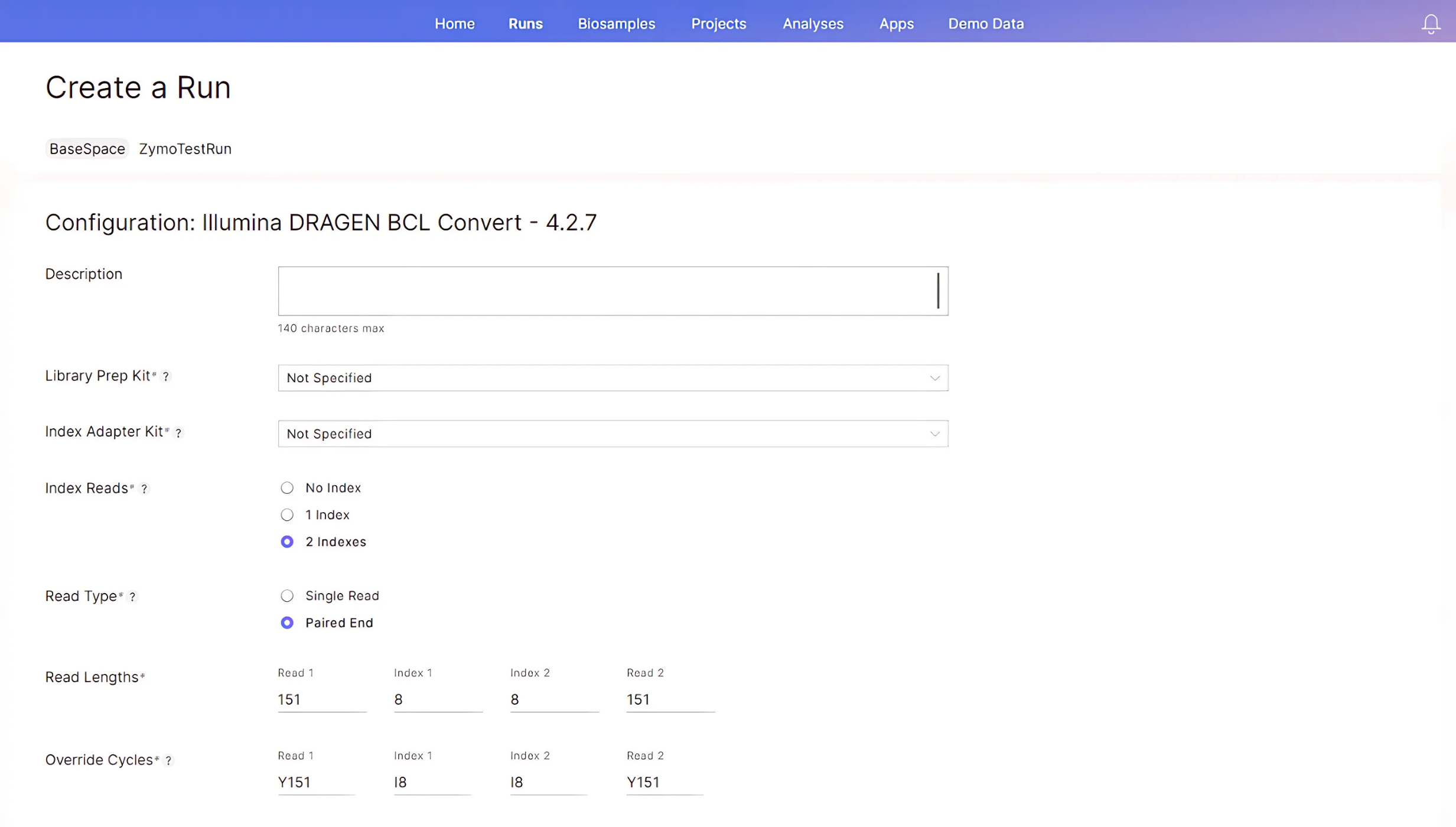
-
In the table below the run parameters, add the number of rows equivalent to the number of samples in the run, then fill out the table with the following sample information:
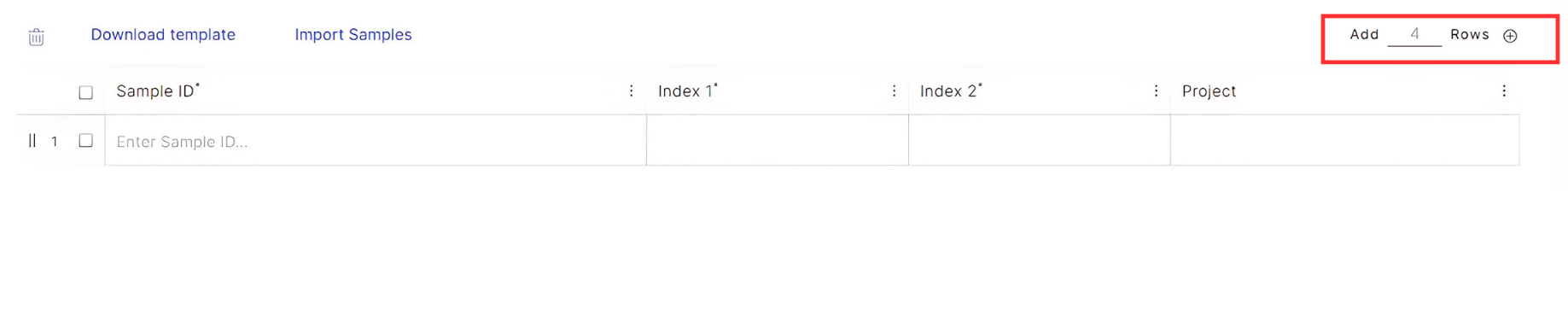
- Sample ID: Input sample IDs of your choice in this column.
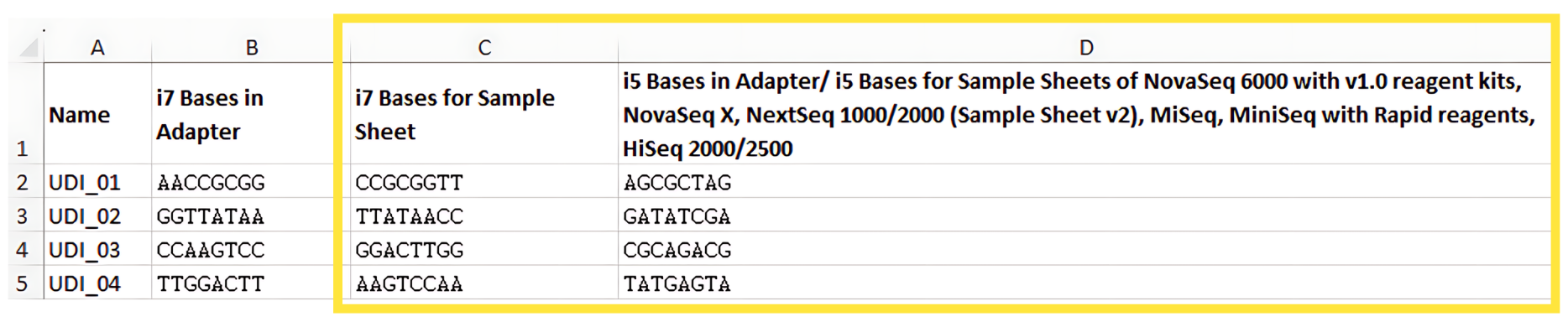
- Index 1: For each sample, copy over the corresponding i7 barcode sequence from column C of the Zymo-Seq UDI Primer Set 1-96 excel sheet labeled “i7 Bases for Sample Sheet”. In this example, samples ZymoSeqRF1-ZymoSeqRF4 have been assigned UDI_01 – UDI_04, respectively.
-
Index 2: For each sample, copy over the corresponding i5 barcode sequence from column D of the Zymo-Seq UDI Primer Set 1-96 excel sheet labeled “i5 Bases in Adapter…”:
-
Optional: Input a project name of your choice in the “Project” column for each sample
-
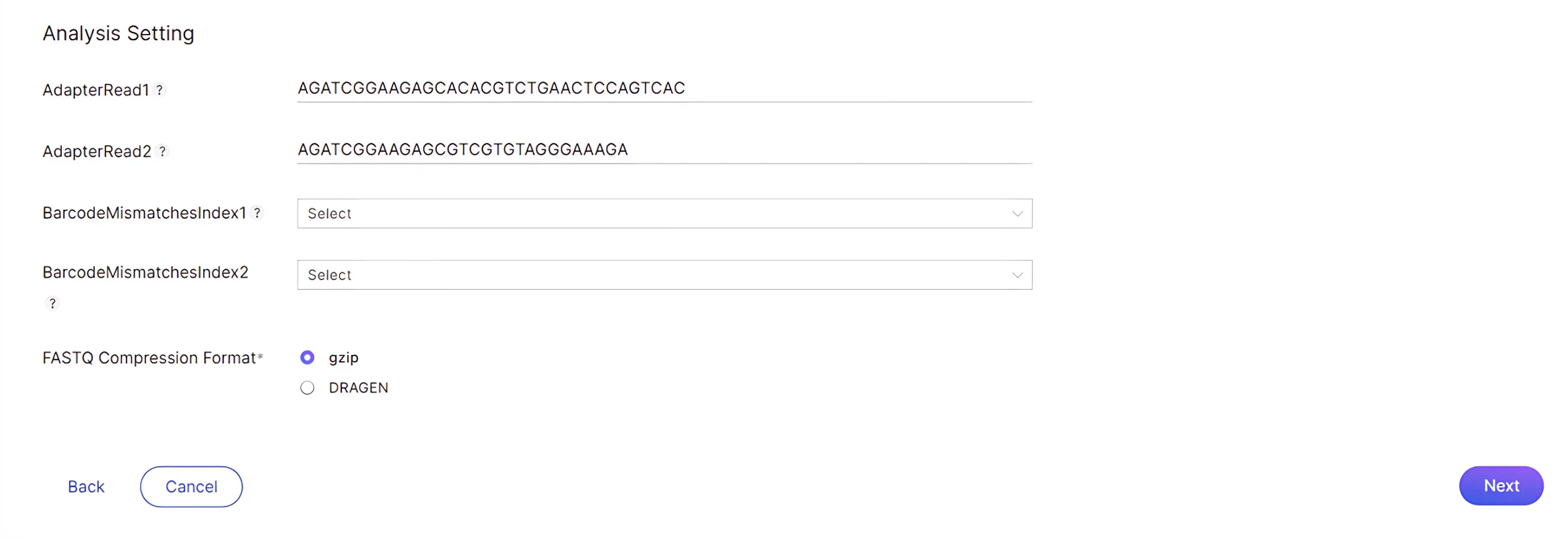
Analysis Settings: Input the adapter sequence to trim for read 1 and read 2. The sequence can be found in Appendix F of the Zymo-Seq RiboFree Total RNA Library Kit protocol.
- AdapterRead1: AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC
- AdapterRead2: AGATCGGAAGAGCGTCGTGTAGGGAAAGA
- Barcode mismatches: This is optional; please indicate the allowed number of index read mismatches. The default is 1 and the maximum value is 2.
-
Select your desired FASTQ Compression format: gzip or DRAGEN:
- Click “Next” to proceed to the “Run Review” Page.
-
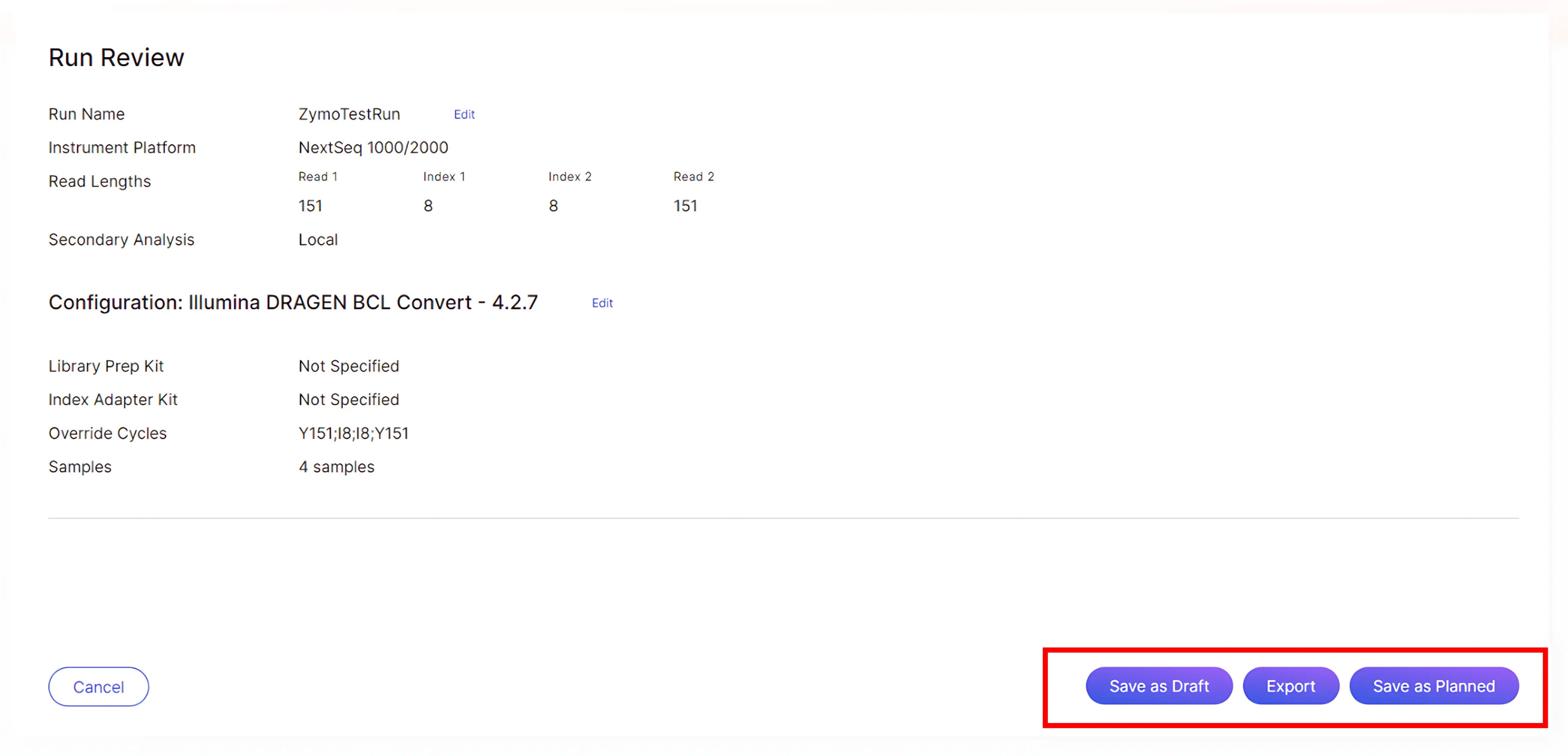
Review and complete the run setup:
- Save as Draft: To save the run setup as a draft for future editing, click “Save as Draft”. Draft runs will be saved in the planned runs list on BaseSpace and will not be available on the NextSeq 2000 for run setup until finalized.
- Export: To save the run setup as a sample sheet in v2 file format, click “Export”. This exported sample sheet can be used to start runs on the NextSeq 2000 in “Local” or “Standalone” mode. If this option is chosen, only local analysis can be done for the run.
- Save as Planned: To finalize and send the run setup to your BaseSpace account, click “Save as Planned”. Runs saved this way will appear in the planned runs list and can be accessed during the NextSeq 2000 run setup in Cloud or Hybrid mode.
Quality NGS libraries generate quality sequencing results, and appropriate software setup as summarized in the steps above ensures streamlined and accurate demultiplexing for smooth downstream analyses. If all these steps are carried out and issues still arise with the run to sequence Zymo-Seq RiboFree Total RNA libraries, Zymo-Seq ATAC libraries, or other versions of libraries, please contact Zymo Research Technical Support.
Learn more about the innovative NGS library prep solutions from Zymo Research