Eliminate Bias in Your Workflow
Use the ZymoBIOMICS® Oral Microbiome Standard to assess potential biases in your workflow, with oral microbiome-specific microorganisms.
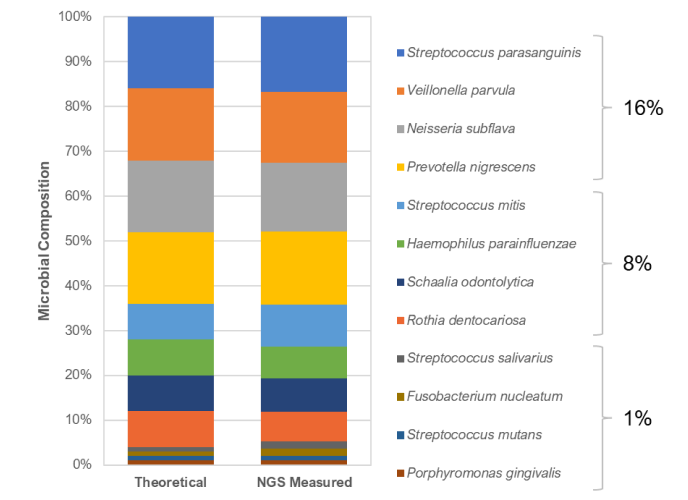
Figure 1. The ZymoBIOMICS® Oral Microbiome Standard is accurately quantified to closely match the theoretical profile by shotgun metagenomic analysis.
True-to-Life Comparison
Designed using data from real, human oral microbiome samples to emulate a representative oral microbiome profile as compared to the Human Oral Microbiome Database (HOMD).
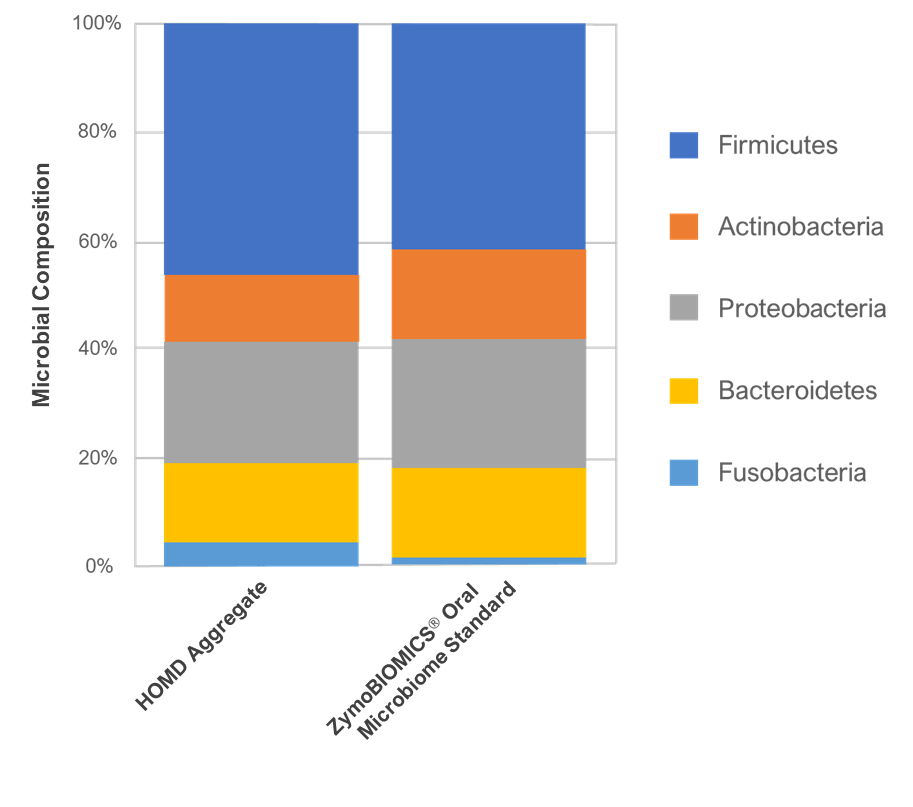
Figure 2. The ZymoBIOMICS® Oral Microbiome Standard accurately represents real, oral microbiome samples at phyla-level.
#1 Cited for a Reason
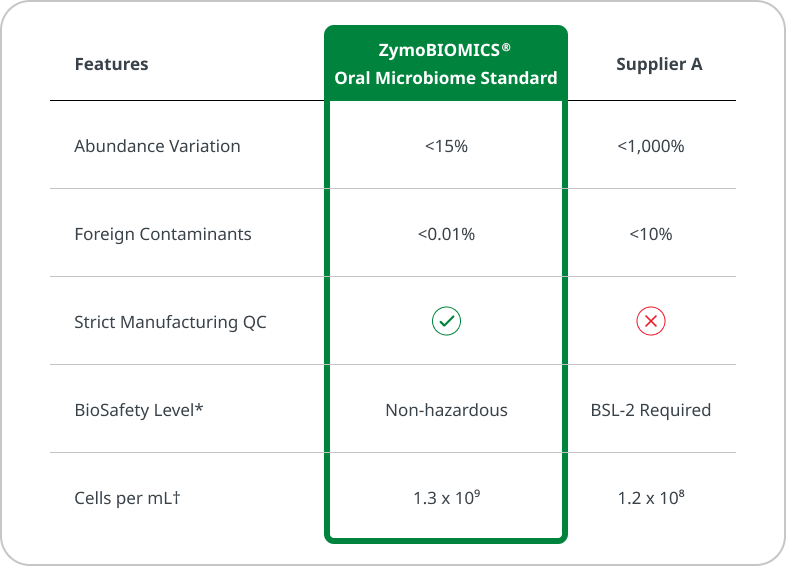
The ZymoBIOMICS® Oral Microbiome Standard offers several advantages over competitors’ standards, providing the best tools for benchmarking your microbiome research.
Next-generation sequencing-powered microbial composition profiling techniques are increasingly common in microbiomics and metagenomics studies. These techniques are known to encounter bias and errors at each workflow step, including DNA extraction, library preparation, sequencing, and bioinformatics analysis. To evaluate different microbiomics workflows, there is a pressing need in the field for reliable reference materials, such as a mock microbial community with a defined composition.
The ZymoBIOMICS® Oral Microbiome Standard comprises 12 bacterial strains with varied abundances to simulate an authentic oral microbiome. It poses challenges for NGS pipelines, including testing lysis efficiency on tough-to-lyse Gram-positive bacteria like Streptococcus mitis, assessing sequencing coverage bias with genomes featuring diverse genomic G/C content, determining detection limits with low-abundance pathogenic organisms, and testing taxonomic resolution with four Streptococcus species. These challenges serve to reveal potential bias in microbiomics or metagenomics workflows. The standard, functioning as a precisely defined input, aids in constructing and optimizing workflows or serves as a quality control tool for routine testing or inter-lab studies.
The microbial standard is accurately characterized, created by combining cells from pure cultures of the 12 microbial strains. Quantification of cells from each pure culture preceded pooling. Post-mixing, NGS-based sequencing confirmed the microbial composition and the absence of impurities (<0.01%).