The Lysis Bias Crisis
Breaking Bug
Liberating DNA from microbial cells can be very difficult. For example, some bacterial cell types are refractory to chemical lysis for various reasons (cell wall thickness, complex lipids, polysaccharides, etc.). Additionally, other microbes such as the opportunistic human pathogen, Cryptococcus neoformans, is notoriously difficult to lyse and has even shown the ability to grow a secondary cell wall in response to chemical lysis techniques [1]. Furthermore, complex microbial communities are very likely to contain similarly tough-to-lyse species, and an accurate profile characterization is impossible if these recalcitrant microbes are not lysed with the same efficiency as the easy-to-lyse species.
Overrepresentation Consternation
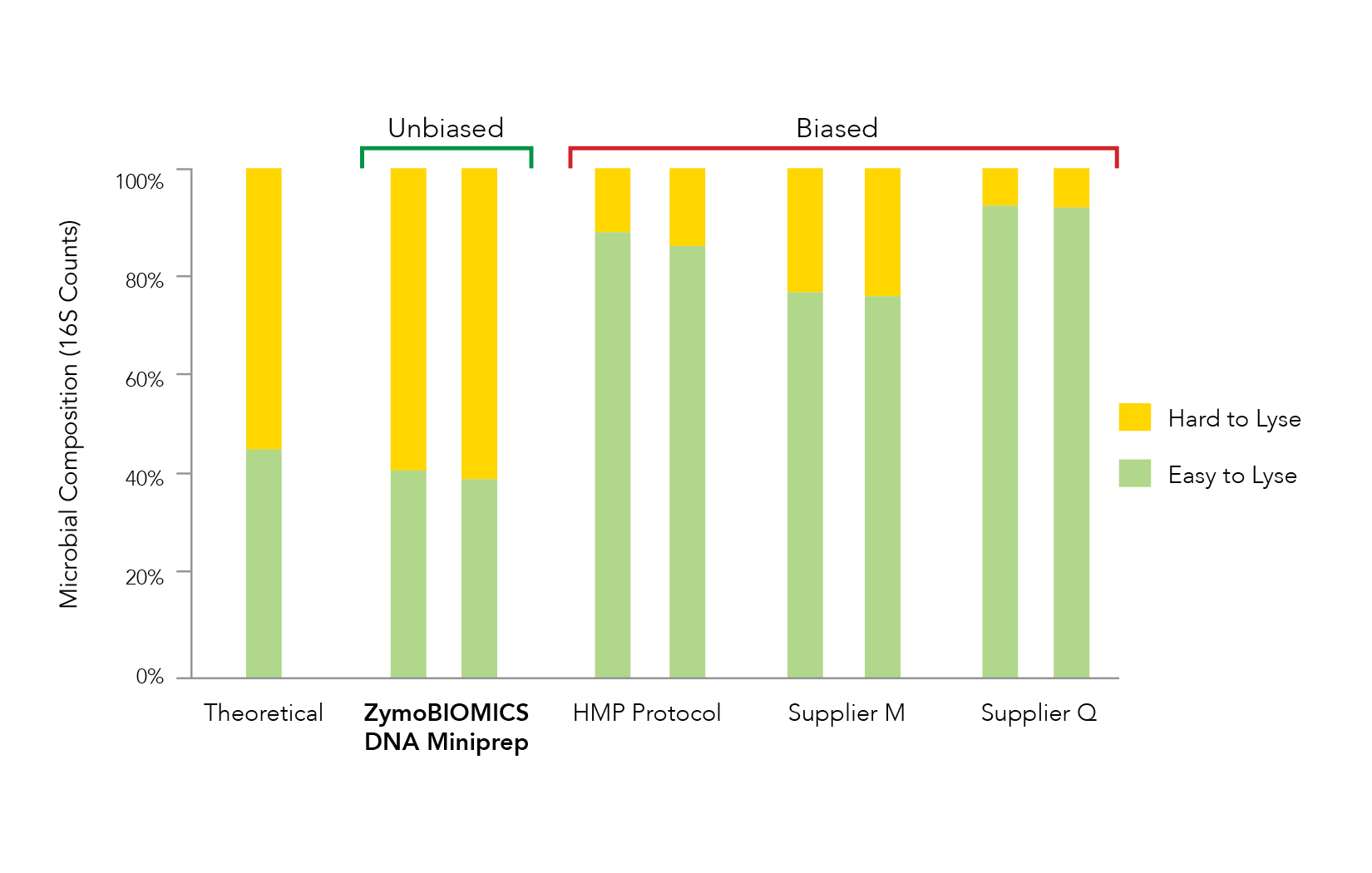
Accurate microbiome profiling requires liberation of DNA from all the diverse species within a microbial community. Unfortunately, it is common to observe ineffective lysis during nucleic acid extraction which leads to microbial profile bias (Figure 1). With more scientists becoming aware of the threat that bias poses for microbiomics, many different cell lysis methods are being investigated and optimized. These include mechanical, chemical, thermal, and enzymatic techniques. While all methods have drawbacks, optimization of all requires strict benchmarking against defined microbial community standards to ensure accurate representation of both easy-to-lyse and tough-to-lyse microbes.
It has been shown that processes utilizing chemical or thermal lysis often cause overrepresentation of easy-to-lyse organisms (e.g. Gram-negative bacteria) due to poor liberation of DNA from tough-to-lyse organisms (e.g. Gram-positive bacteria and yeast) [2-3]. Additionally, enzymatic lysis is inherently non-stochastic by nature and consequently is vulnerable to biases, especially from highly diverse sample types such as soil.
Deus Ex Mechanical
Mechanical lysis methodologies (e.g. sonication, blending, liquid nitrogen/mortar and pestle, French pressing, and bead beating) are considered the best approaches for microbial lysis due to their stochastic nature, with bead beating often referred to as the gold standard. However, if not optimized, mechanical lysis methods can suffer from problems such as low yields, excessive nucleic acid shearing, non-uniform lysis due to incomplete lysis, excessive heat and shear forces, and inefficient lysis of tough-to-lyse microbes, respectively. If a bead beating protocol is not validated to overcome these issues, the resulting microbial profile may be biased. Unfortunately, simply combining an array of cell lysis mechanisms does not necessarily reduce bias, despite potentially improving yields.
Dedicated to accuracy
To help all microbiome researchers produce the most accurate microbial profiling data, Zymo Research has evaluated several bead beating procedures and determined the best parameters to yield unbiased nucleic acid extractions. The use of the ZymoBIOMICS Microbial Community Standard and the only validated, bias-free DNA extraction kit, the ZymoBIOMICS DNA Miniprep Kit, made it possible to benchmark these methods. Zymo Research is passionate about making sure the microbiomics field has the best research tools to create accurate and reproducible data. Below we have provided beat beating protocols that have been extensively tested using the ZymoBIOMICS Microbial Community Standard and validated as unbiased using Next-Gen Sequencing.
With influences in human health, agriculture, and environmental safety, it is critical for microbiomics studies to generate accurate and reproducible data. Poor inter-lab reproducibility due to biased lysis techniques has plagued the field of microbiomics but, we are at a turning point where scientists are seeking benchmarked and standardized methods. By utilizing validated lysis parameters, researchers can have more confidence in the accuracy of their microbial profiling data.
Validated Bead Beating Protocols using the ZymoBIOMICS DNA Miniprep kit:
MP Fastprep-24 Bead Beating Protocol:
(with 2 ml BashingBead Tubes)
Maximum of 20 tubes. The weight of > 20 tubes may cause a system error.
- 1 minute on at max speed
- 5 minutes rest
- Repeat cycle 5 times for a total of 5 minutes of bead beating
Biospec Mini-BeadBeater-96 Bead Beating Protocol:
(with 2 ml BashingBead Tubes)
- 5 minutes on at Max RPM
- 5 minutes rest
- Repeat cycle 4 times for a total of 20 minutes of bead beating
Biospec Mini-BeadBeater-96 Bead Beating Protocol:
(with 96 well lysis rack)
- 5 minutes on Max RPM
- 5 minutes rest
- Repeat cycle 8 times for a total of 40 minutes of bead beating
Bertin Precelys Evolution Bead Beating Protocol:
(with 2 ml BashingBead Tubes)
- 1 minute on at 9,000 RPM
- 2 minutes rest
- Repeat cycle 4 times for a total of 4 minutes of bead beating
Vortex Genie Bead Beating Protocol:
(with Horizontal-(24) Microtube Adaptor)
Maximum of 18 tubes. The weight of > 18 tubes will slow the mechanism and introduce the bias.
- 40 minutes of continuous bead beating (max of 18 tubes per adaptor)
References:
1. Farkaš V, Takeo K, Maceková D, Ohkusu M, Yoshida S, Sipiczki M. Secondary cell wall formation in Cryptococcus neoformans as a rescue mechanism against acid-induced autolysis. FEMS Yeast Research, 2009, 9(2): 311-320
2. Sohrabi M, Nair RG, Samaranayake LP, Zhang L, Zulfiker AH, Ahmetagic A, Good D, Wei MQ. The yield and quality of cellular and bacterial DNA extracts from Human oral rinse samples are variably affected by the cell lysis methodology. Journal of Microbiological Methods 2016, 122: 64-72
3. Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of Methods for the Extraction and Purification of DNA from the Human Microbiome. PLOS ONE 2012.


