Microbiome-Grade DNA Extraction Kits
New requirements have made old methods obsolete.
Many publications have reported dramatic variations in microbiome profiling due to the use of different DNA extraction methods [1-6]. Biased lysis during DNA extraction is likely the biggest factor causing poor data reproducibility in these studies. Many of the DNA extraction methods used in microbiome profiling were developed before this field took off and unfortunately, they were not designed to meet microbiomics’ rigorous demands: being unbiased and having low background contamination.
The most cited (popular) methods are dated.
The three most cited methods for DNA extraction in microbiomics publications preferentially lyse specific classes of microbes (Figure 1). These include the fecal DNA extraction protocol used and recommended by the Human Microbiome Project (HMP). This method and others, when used for metagenomic profiling, dramatically over-represent the abundance of easy-to-lyse microbes, (e.g. Gram-negative microbes) as indicated in Figure 1. When it comes to fecal samples, these methods tend to over-represent the abundance of Bacteroidetes and Proteobacteria and underestimate the abundance of Firmicutes and Actinobacteria (Figure 2). This most likely explains the contradictory interpretations of the human gut microbiome by HMP and MetaHIT, which was described in a previous article.
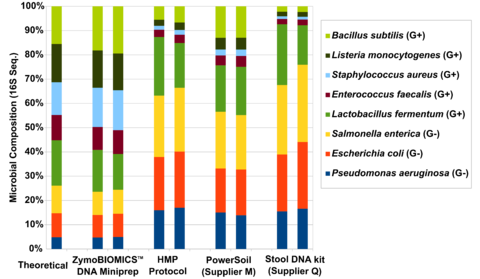
Figure 1. Assessing the performance of four different DNA extraction kits with the ZymoBIOMICS™ Microbial Community Standard . The four different DNA extraction methods investigated include ZymoBIOMICS™ DNA Miniprep Kit , Human Microbiome Project fecal DNA extraction protocol (HMP Protocol), a soil DNA extraction kit from “Supplier M” and a fecal DNA extraction kit from Supplier Q. Extracted DNA was subjected to 16S sequencing with an internal library preparation protocol. The microbial composition was determined by mapping raw sequencing reads against reference 16S sequences of the strains contained in the standard.
Why is the HMP fecal DNA extraction protocol biased?
The HMP protocol lyses microbes thermally (95°C incubation), chemically (SDS in lysis buffer), and mechanically (vortexing with 0.7 mm Garnet beads). Unfortunately, using more cell lysis mechanisms does not necessarily result in less bias. Thermal and chemical methods are very effective in disrupting easy-to-lyse, Gram-negative microbes (e.g. Bacteroidetes and Proteobacteria), but not tougher Gram-positive organisms (e.g. yeasts and spores). Although mechanical lysis is generally regarded as less biased due to its stochastic nature, we have found that the size and density of the beads play a critical role in bead driven lysis. What was found in the case of the HMP protocol is that the 0.7 mm Garnet beads are too large to effectively lyse small bacteria and therefore underrepresent the “tough-to-lyse” population (i.e. Firmicutes). Compounding the issue further, the thermal and chemical methods effectively lyse the gram-negative organisms and boost their population in the final profile, amplifying the bias.
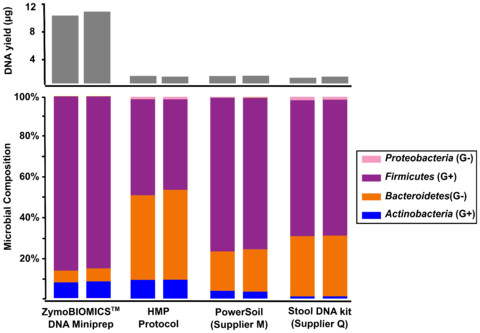
Figure 2. Comparing four different DNA extraction kits with one fecal sample. The four different DNA extraction methods investigated include ZymoBIOMICS™ DNA Miniprep Kit, Human Microbiome Project fecal DNA extraction protocol (HMP Protocol), a soil DNA extraction kit from “Supplier M” and a fecal DNA extraction kit from “Supplier Q.” Extracted DNA was subjected to 16S sequencing with an internal library preparation protocol. The microbial composition was determined using Qiime 1.9.1.
Extraction Kits Designed and Validated for Microbiome Measurements.
The first step in designing an extraction kit for microbiome measurements is benchmarking the performance of the technology against a known input (e.g. a Microbiome Community Standard). With a diverse whole-cell standard, Zymo Research was able to develop a technology that achieved complete lysis of a range of organisms comprising different sizes and cell wall recalcitrance. What we found was that a single bead type was incapable of achieving complete (or statistically close enough) lysis of every microbe tested. Instead, we found that a formulation of 0.1 mm and 0.5 mm ultra-high density beads and a unique lysis buffer ensured complete microbial cell disruption regardless of cell wall hardiness and size (Figure 1). With this enhanced lysis system, we see the same fecal sample in a completely different light, where Firmicutes actually dominate the population (Figure 2). It can be reasoned that this is the true profile (or closest to the truth) because we have evidence of unbiased lysis in the Microbiome Standard while the three most cited methods are poorly lysing the tougher organisms and thus greatly over representing the easy-to-lyse organisms (Figure 1). This supposition is further supported by the fact that the yield for the fecal sample is substantially higher (Figure 2) reflecting the greater ability to lyse the tough organisms (Firmicutes and Actinobacteria) which represent a majority population within the stool sample tested.
Try a free sample of the ZymoBIOMICS DNA Miniprep Kit for unbiased sample lysis:
Get Free SampleReferences:
1. Mitchell KR, Takacs-Vesbach CD: A comparison of methods for total community DNA preservation and extraction from various thermal environments. Journal of industrial microbiology & biotechnology 2008, 35(10):1139-1147.
2. Vishnivetskaya TA, Layton AC, Lau MC, Chauhan A, Cheng KR, Meyers AJ, Murphy JR, Rogers AW, Saarunya GS, Williams DE et al: Commercial DNA extraction kits impact observed microbial community composition in permafrost samples. FEMS microbiology ecology 2014, 87(1):217-230.
3. Hart ML, Meyer A, Johnson PJ, Ericsson AC: Comparative Evaluation of DNA Extraction Methods from Feces of Multiple Host Species for Downstream Next-Generation Sequencing. PloS one 2015, 10(11):e0143334.
4. Kennedy NA, Walker AW, Berry SH, Duncan SH, Farquarson FM, Louis P, Thomson JM, Satsangi J, Flint HJ, Parkhill J et al: The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PloS one 2014, 9(2):e88982.
5. Sohrabi M, Nair RG, Samaranayake LP, Zhang L, Zulfiker AH, Ahmetagic A, Good D, Wei MQ: The yield and quality of cellular and bacterial DNA extracts from human oral rinse samples are variably affected by the cell lysis methodology. Journal of microbiological methods 2016, 122:64-72.
6. Gerasimidis K, Bertz M, Quince C, Brunner K, Bruce A, Combet E, Calus S, Loman N, Ijaz UZ: The effect of DNA extraction methodology on gut microbiota research applications. BMC research notes 2016, 9:365.


