The Missing Microbial Ingredient
Until recently, the area of microbiomics and metagenomics has been relatively unrecognized as a major field in biological research with only a few dozen publications a year. However, with the advent of Next-Generation sequencing (NGS) and the ability to sequence millions of mixed DNA sequences simultaneously, microbiome and metagenomic studies have expanded in to nearly every area of biological research today including patient care. Although NGS has opened the door to this data rich field, it is recognized by most experts that significant technical advancements will be required to produce accurate and valid data sets moving forward.
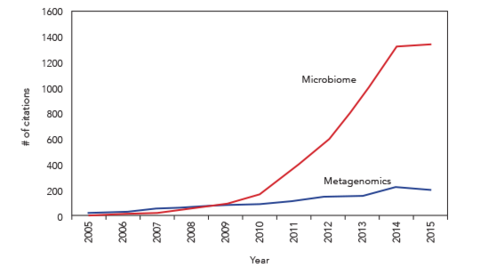
Because of this rapid expansion and the demand for high-performance protocols at almost every level of sample processing, specially designed controls and reagents are needed, including sample collection, DNA and RNA extraction, NGS library preparation, and special bioinformatic software to understand the variations throughout these many steps. It is clear that both whole cell microbial reference standards as well as genomic DNA standards are required to ascertain detection limits and performance statistics with all studies including those clinical samples.Implementing microbial reference controls into microbiome studies is a new required practice to ensure high-quality data. However, fabrication of high-quality reference controls has been difficult with the only source being the well-known BEI control DNA controls, which are now in very limited supply. Additionally, since efficient microbial lysis is a paramount step in all microbiome studies, the need for accurately quantified DNA and cellular mixed microbial standards is also needed to determine detection limits and percent recovery of various types of organisms. Recognizing these needs, three groups addressed the challenge and fabricated multiple microbial reference standards, including Zymo Research (ZymoBIOMICS®), the Association of Biomolecular Resources Facilities (ABRF) metagenomics research group (Class I MRGR standards) and the National Institute of Standards and Testing (NIST). While only a few of these whole cell and genomics standards are currently available, future standards including complex mixtures of both eukaryotic and prokaryotic as well as RNA are currently being fabricated.
The use of microbial reference standards is an important ingredient to be considered for any microbiome project
The ZymoBIOMICS® Microbial Community Standard is a mock microbial community consisting of 8 bacterial and 2 fungal strains (3 Gram-negative, 5 Gram-positive and 2 yeasts) with 7 being human pathogens. In contrast, the ABRF MGRG controls include 10 strains of belonging to Class I genomes, and include both Gram negative and positive and 1 archaea but does not include human pathogen related strains. NIST has generated several microbial standards, with the most recent being a human microbiome-related panel of microbial DNA with a release date in late 2017. Both Zymo Research and the ABRF offer a whole cell microbial standard which are absolutely necessary for determining DNA extraction efficiency or as use as sample/matrix spike-in for recovery determinations, which is certainly one of the largest shortcoming of most DNA kits today.
Regardless of the study, whether it be clinical FMT samples, routine metagenomic samples from soil or food, or DNA extraction efficiency studies, the use of microbial reference standards is an important ingredient to be considered for any microbiome project as it will enable biologists, software developers, and product manufactures to determine efficiencies at every point in their process.


