Addition by Subtraction - Host DNA Depletion for Microbiome Analyses
Host DNA Depletion: Making the Most of WGS
Next-Gen Sequencing, once considered a luxury service that cost hundreds of thousands of dollars and countless hours of time, is rapidly becoming the go-to technology for sample identification thanks to incredible improvements in cost and efficiency.
As the field of microbiomics has continued to grow, so has the appeal of whole genome sequencing (WGS), which utilizes parallel sequencing of genome fragments to rapidly determine the complete DNA sequence of an organism. When applied to a microbiome sample, WGS can identify strain-level differences among microbial species. Additionally, the cost of sequencing has dropped tremendously, from the $2.7 billion Human Genome Project to the $1,000 genome of today. Because of its increased accessibility, comprehensive coverage of organisms present in a sample, and ability to identify novel genomes, WGS is now a realistic and desirable option for microbial sample identification.
The Host with the Most (Contaminating DNA)
An increasingly important application of microbiomics is how the microbes living in and on humans affect us, for better or worse. A major challenge to assessing the human microbiome with shotgun sequencing is the presence of human host DNA that “contaminates” the microbiome sample.
In clinical samples, such as skin swabs or biological fluids, WGS results are dominated by sequences from the human genome. Even though the expense of sequencing has decreased significantly, this type of host contamination negates some of this benefit by diminishing the amount of relevant data produced. For this reason, a method to remove the host DNA prior to sequencing is essential to maximizing relevant data while leveraging the power of WGS.
Addition by Subtraction
To overcome the challenge of contaminating host DNA, Zymo Research has reimagined the DNA isolation process with the HostZERO Microbial DNA Kit. This host DNA depletion kit uses a novel method to reduce the amount of contaminating host DNA by selectively lysing the human cells and degrading this DNA prior to total DNA purification. Paired with the most accurate purification technology available, the HostZERO Microbial DNA Kit ensures the exclusive capture of DNA from living microbial cells in a biological sample.
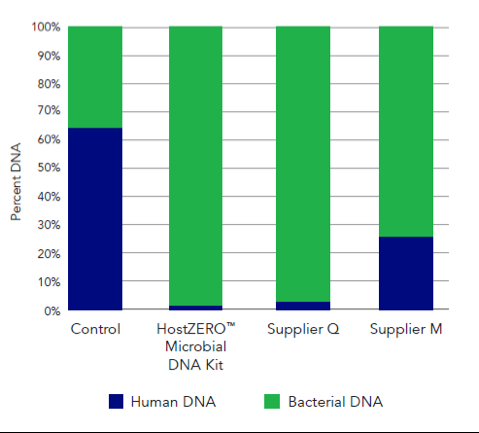
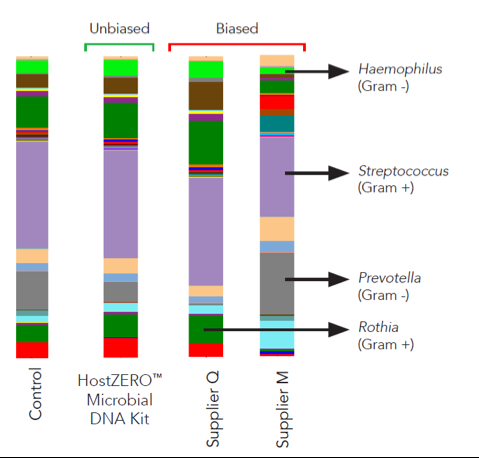
This new technology reduces the presence of human DNA in a saliva sample from 64.5% in the untreated sample to just 0.8% in the treated sample (Figure 1). Additionally, the HostZERO technology purifies the retained bacterial DNA without bias toward easy-to-lyse cells (Figure 2). With the removal of host DNA and reduction of bias in purification, the HostZERO Microbial DNA Kit boosts the power of WGS by allowing more sequencing reads to be focused on microbial DNA.
Making the Most of the WGS Situation
As the price of WGS continues to fall, more and more microbiome profiling projects are turning to metagenomic sequencing to reveal the microbial species within environmental and even clinical samples. But, without additional sample processing, human-derived DNA samples can contain up to twice as much DNA from the host genome than from the sample’s microbiome, which wastes valuable sequencing information. The innovative HostZERO Microbial DNA Kit effectively depletes host DNA while maintaining the microbial profile of biological samples. Pairing the host DNA depletion of HostZERO with the vast data-producing power of WGS generates the highest quality and highest volume metagenomic data for biological samples.


